Mapping open chromatin by ATAC-seq in bread wheat.
Wang X, Chen C, He C, Chen D, Yan W
Front Plant Sci. 2022 Nov 16;13:1074873. doi: 10.3389/fpls.2022.1074873.
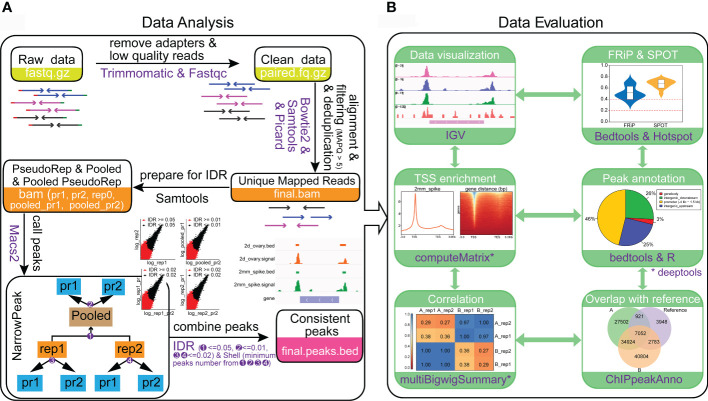
Gene transcription is largely regulated by cis-regulatory elements. Assay forTransposase-Accessible Chromatin using sequencing (ATAC-seq) is an emergingtechnology that can accurately map cis-regulatory elements in animals andplants. However, the presence of cell walls and chloroplasts in plants hindersthe extraction of high-quality nuclei, thereby affects the quality of ATAC-seqdata. Meanwhile, it is tricky to perform ATAC-seq with different tissue types,especially for those with limited size and amount. Moreover, with rapid growthof ATAC-seq datasets from plants, powerful and easy-to-use data analysispipelines for ATAC-seq, especially for wheat is lacking. Here, we provided anall-in-one solution for mapping open chromatin in wheat including bothexperimental and data analysis procedure. We efficiently obtained nuclei withless cell debris from various wheat tissues. High-quality ATAC-seq data fromyoung spike and ovary, which are hard to harvest were generated. We determinedthat the saturation sequencing depth of wheat ATAC-seq is about 16 Gb.Particularly, we developed a powerful and easy-to-use online pipeline to analyzethe wheat ATAC-seq data and this pipeline can be easily extended to other plantspecies. The method developed here will facilitate plant regulatory genome studynot only for wheat but also for other plant species.