TripletCell: a deep metric learning framework for accurate annotation of celltypes at the single-cell level.
Liu Y, Wei G, Li C, Shen LC, Gasser RB, Song J, Chen D*,Yu DJ
Brief Bioinform. 2023 May 19;24(3):bbad132. doi: 10.1093/bib/bbad132.
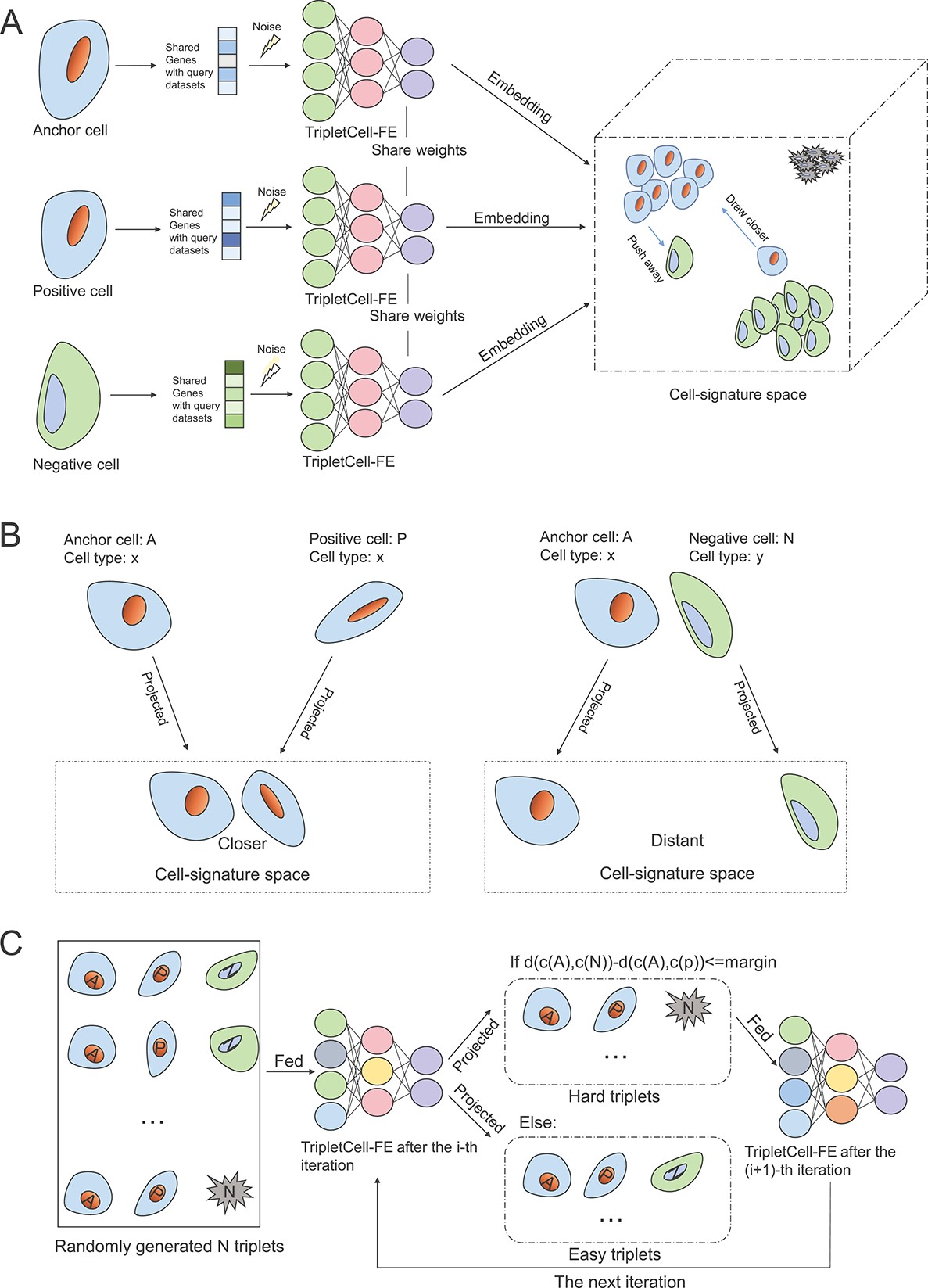
Single-cell RNA sequencing (scRNA-seq) has significantly accelerated theexperimental characterization of distinct cell lineages and types in complextissues and organisms. Cell-type annotation is of great importance in most ofthe scRNA-seq analysis pipelines. However, manual cell-type annotation heavilyrelies on the quality of scRNA-seq data and marker genes, and therefore can belaborious and time-consuming. Furthermore, the heterogeneity of scRNA-seqdatasets poses another challenge for accurate cell-type annotation, such as thebatch effect induced by different scRNA-seq protocols and samples. To overcomethese limitations, here we propose a novel pipeline, termed TripletCell, forcross-species, cross-protocol and cross-sample cell-type annotation. Wedeveloped a cell embedding and dimension-reduction module for the featureextraction (FE) in TripletCell, namely TripletCell-FE, to leverage the deepmetric learning-based algorithm for the relationships between the reference geneexpression matrix and the query cells. Our experimental studies on 21 datasets(covering nine scRNA-seq protocols, two species and three tissues) demonstratethat TripletCell outperformed state-of-the-art approaches for cell-typeannotation. More importantly, regardless of protocols or species, TripletCellcan deliver outstanding and robust performance in annotating different types ofcells. TripletCell is freely available at https://github.com/liuyan3056/TripletCell. We believe that TripletCell is areliable computational tool for accurately annotating various cell types usingscRNA-seq data and will be instrumental in assisting the generation of novelbiological hypotheses in cell biology.