Small RNAs in angiosperms: sequence characteristics, distribution and generation.
Chen D, Meng Y, Ma X, Mao C, Bai Y, Cao J, Gu H, Wu P, Chen M
Bioinformatics. 2010 Jun 1;26(11):1391-4. doi: 10.1093/bioinformatics/btq150.
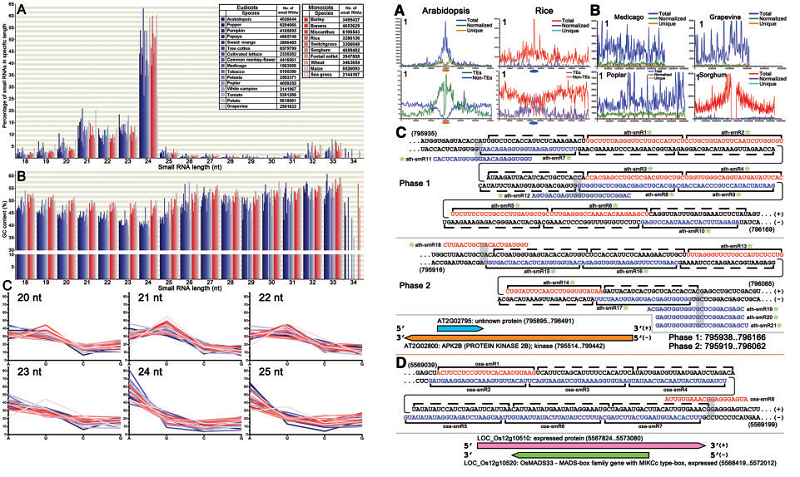
High-throughput sequencing (HTS) has opened up a new era for small RNA (sRNA) exploration. Using HTS data for a global survey of sRNAs in 26 angiosperms, elevated GC contents were detected in the monocots, whereas the 5(')-terminal compositions were quite uniform among the angiosperms. Chromosome-wide distribution patterns of sRNAs were investigated by using scrolling-window analysis. We performed de novo natural antisense transcript (NAT) prediction, and found that the overlapping regions of trans-NATs, but not cis-NATs, were hotspots for sRNA generation. One cis-NAT generates phased natural antisense short interfering RNAs (nat-siRNAs) specifically from flowers in Arabidopsis, while one in rice produces phased nat-siRNAs from grains, suggesting their organ-specific regulatory roles.