A reference single-cell regulomic and transcriptomic map of cynomolgus monkeys
Qu J, Yang F#, Zhu T#, Wang Y, Fang W, Ding Y, Zhao X, Qi X, Xie Q, Chen M, Xu Q, Xie Y, Sun Y, Chen D*
Nat Commun. 2022 Jul 13;13(1):4069.doi: 10.1038/s41467-022-31770-x.
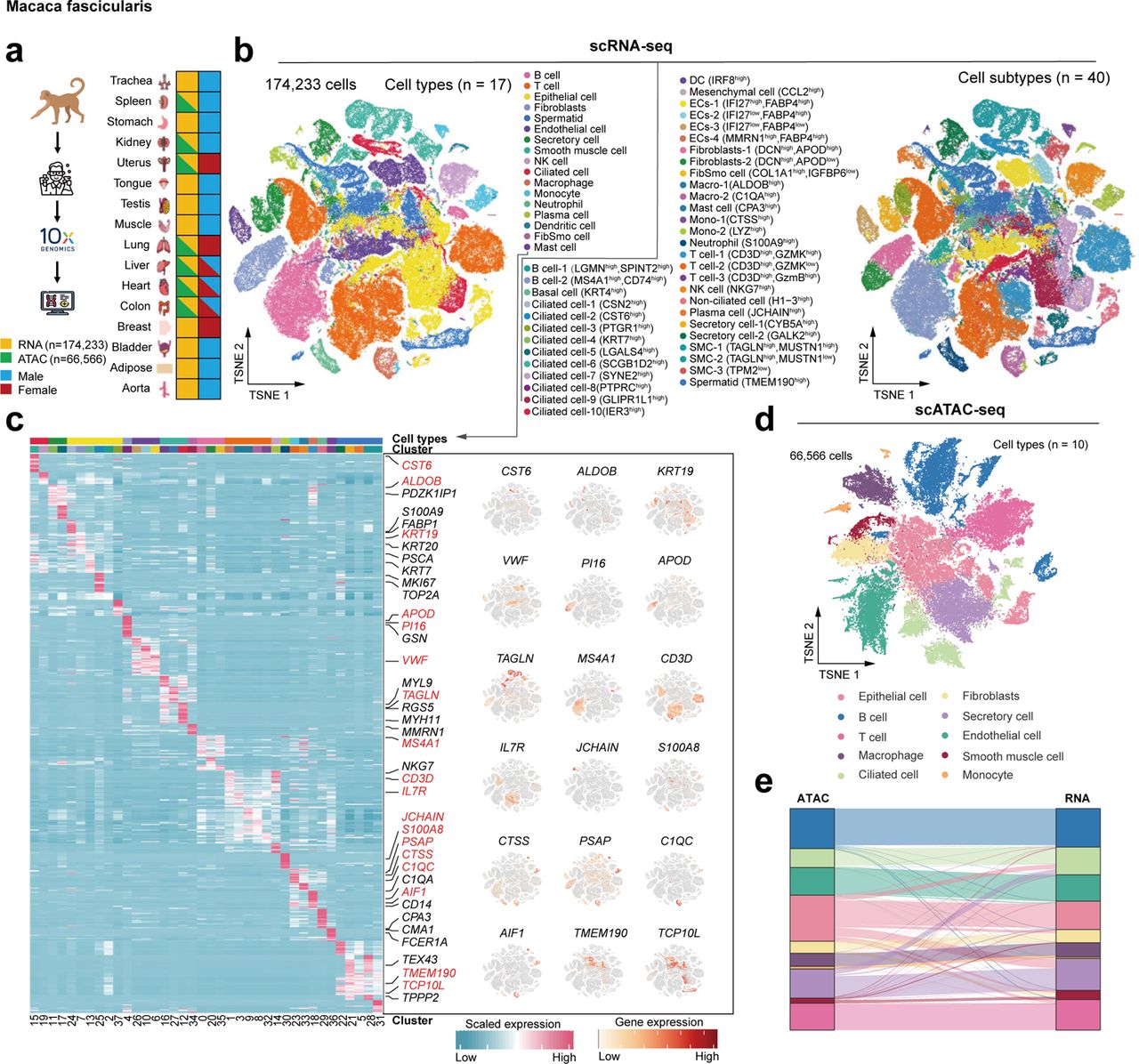
Non-human primates are attractive laboratory animal models that accurately reflect both developmental and pathological features of humans. Here we present a compendium of cell types across multiple organs in cynomolgus monkeys (Macaca fascicularis) using both single-cell chromatin accessibility and RNA sequencing data. The integrated cell map enables in-depth dissection and comparison of molecular dynamics, cell-type compositions and cellular heterogeneity across multiple tissues and organs. Using single-cell transcriptomic data, we infer pseudotime cell trajectories and cell-cell communications to uncover key molecular signatures underlying their cellular processes. Furthermore, we identify various cell-specific cis-regulatory elements and construct organ-specific gene regulatory networks at the single-cell level. Finally, we perform comparative analyses of single-cell landscapes among mouse, monkey and human. We show that cynomolgus monkey has strikingly higher degree of similarities in terms of immune-associated gene expression patterns and cellular communications to human than mouse. Taken together, our study provides a valuable resource for non-human primate cell biology.