An angiosperm NLR Atlas reveals that NLR gene reduction is associated withecological specialization and signal transduction component deletion.
Liu Y, Zeng Z, Zhang YM, Li Q, Jiang XM, Jiang Z, Tang JH,Chen D, Wang Q, Chen JQ, Shao ZQ
Mol Plant. 2021 Dec 6;14(12):2015-2031. doi: 10.1016/j.molp.2021.08.001.
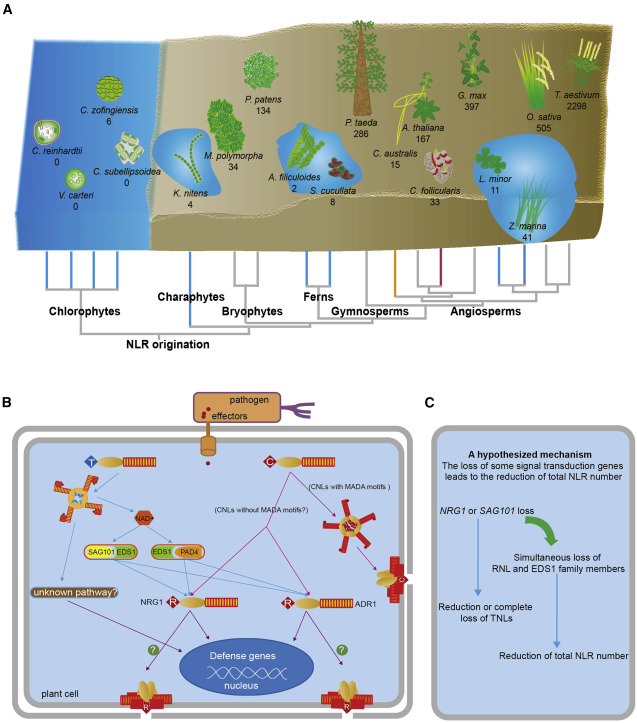
Nucleotide-binding leucine-rich-repeat (NLR) genes comprise the largest familyof plant disease-resistance genes. Angiosperm NLR genes are phylogeneticallydivided into the TNL, CNL, and RNL subclasses. NLR copy numbers and subclasscomposition vary tremendously across angiosperm genomes. However, theevolutionary associations between genomic NLR content and ecological adaptation,or between NLR content and signal transduction components, are poorlycharacterized because of limited genome availability. In this study, weestablished an angiosperm NLR atlas (ANNA, https://biobigdata.nju.edu.cn/ANNA/)that includes NLR genes from over 300 angiosperm genomes. Using ANNA, werevealed that NLR copy numbers differ up to 66-fold among closely relatedspecies owing to rapid gene loss and gain. Interestingly, NLR contraction wasassociated with adaptations to aquatic, parasitic, and carnivorous lifestyles.The convergent NLR reduction in aquatic plants resembles the lack of NLRexpansion during the long-term evolution of green algae before the colonizationof land. A co-evolutionary pattern between NLR subclasses and plant immunepathway components was also identified, suggesting that immune pathwaydeficiencies may drive TNL loss. Finally, we identified a conserved TNL lineagethat may function independently of the EDS1-SAG101-NRG1 module. Collectively,these findings provide new insights into the evolution of NLR genes in thecontext of ecological adaptation and genome content variation.