Systematic annotation of conservation states provides insights into regulatoryregions in rice.
Zhou X#, Zhu T#, Fang W, Yu R, He Z, Chen D*.
J Genet Genomics. 2022 Apr 22:S1673-8527(22)00123-0. doi:10.1016/j.jgg.2022.04.003.
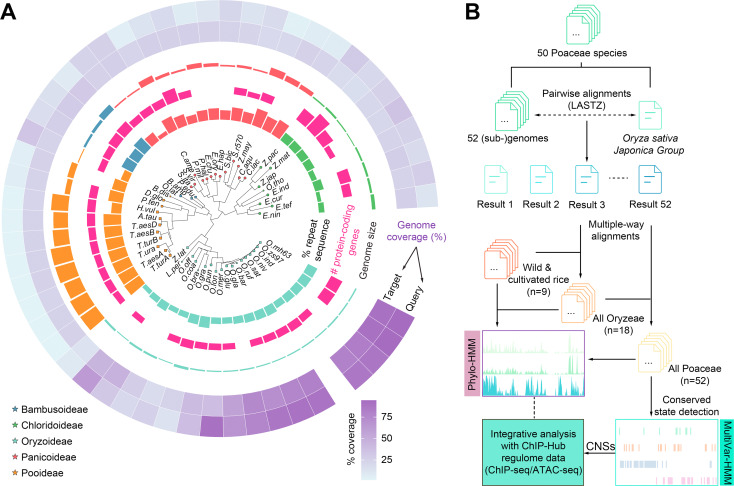
Plant genomes contain a large fraction of non-coding sequences. Discovery andannotation of conserved non-coding sequences (CNSs) in plants is an ongoingchallenge. Here report the we application of comparative genomics tosystematically identify CNSs in 50 well-annotated Gramineae genomes using rice(Oryza sativa) as the reference. We conduct multiple-way whole genome alignmentsto the rice genome. The rice genome is annotated as 20 conservation states (CSs)at single nucleotide resolution using a multivariate hidden Markov model(ConsHMM) based on the multiple-genome alignments. Different states showdistinct enrichments for various genomic features and the conservation scores ofCSs are highly correlated with the level of associated chromatin accessibility.We find that at least 33.5% of the rice genome is highly under selection withmore than 70% of the sequence lying outside of coding regions. A catalog of855,366 regulatory CNSs is generated and they significantly overlapped withputative active regulatory elements such as promoters, enhancers, andtranscription factor binding sites. Collectively, our study provides a resourcefor studying functional non-coding regions of the rice genome and anevolutionary aspect of regulatory sequences in higher plants.